附:在部分配对的数据上训练模型
如果数据中有一部分细胞同时检测的配对多组学细胞,这样的配对信息也可以被利用起来提高整合精度,下面我们将说明如何让GLUE利用配对细胞。
[1]:
from itertools import chain
from random import Random
import anndata as ad
import networkx as nx
import pandas as pd
import scanpy as sc
import scglue
import seaborn as sns
from matplotlib import rcParams
[2]:
scglue.plot.set_publication_params()
rcParams["figure.figsize"] = (4, 4)
读取预处理数据
首先,读取 第一步 产生的预处理数据。
[3]:
rna = ad.read_h5ad("rna-pp.h5ad")
atac = ad.read_h5ad("atac-pp.h5ad")
guidance = nx.read_graphml("guidance.graphml.gz")
使用 obs_names 标记配对的细胞
在 scglue 中,我们用AnnData对象的 obs_names 来识别不同模态中配对的细胞。具体来说,在不同模态里具有相同的 obs_name 的细胞被视为配对的,而具有不同的 obs_name 的细胞被视为不配对的。
The dataset we are using is generated by SNARE-seq, which produces paired scRNA-seq and scATAC-seq profiles. In previous tutorials, we intentionally anonymized the paired cells by appending modality name to the cell barcodes (“_RNA”, “_ATAC”) to mimic an unpaired dataset:
[4]:
rna.obs_names, atac.obs_names
[4]:
(Index(['09L_GTTAGAACAGAT_RNA', '09I_CATAGCTCAACG_RNA', '09G_TTACAGGCAGGT_RNA',
'09B_CTATGGGCGTGC_RNA', '09L_AGTGATTGTTCG_RNA', '09C_ATATGTCTATAA_RNA',
'09A_CCTATTATTCGA_RNA', '09C_GCGTGCCGAATG_RNA', '09G_AGCTGACGCCGA_RNA',
'09D_CAGTCCCCAAAT_RNA',
...
'09H_GTTCGCTTGTCG_RNA', '09B_CAGGTGGTCTAT_RNA', '09C_GACCCTGTCGTT_RNA',
'09I_AACCAATCCTCA_RNA', '09C_AGAGTCTGAAAC_RNA', '09E_CCTGCTCGTCAC_RNA',
'09E_TGAGCACCTCCT_RNA', '09E_CCGGACTCGATC_RNA', '09F_GCCCGACTCGCA_RNA',
'09D_ATACTGCGCCCA_RNA'],
dtype='object', name='cells', length=9190),
Index(['09L_GTAAACCCGATA_ATAC', '09D_GCCGGGCGGCCG_ATAC',
'09J_TAGGCCTGAATT_ATAC', '09C_TCCGGCATCTCA_ATAC',
'09C_AGGCCACGTACA_ATAC', '09E_AAGGGGAGCTGG_ATAC',
'09K_CCGCCTTATAAT_ATAC', '09A_CGGATGTATCAT_ATAC',
'09B_TCAGAACTGCTA_ATAC', '09J_TCCGTGATAGAG_ATAC',
...
'09E_TAGTCGCGTACT_ATAC', '09J_GCACCTAAATGA_ATAC',
'09E_CACCCACGCTCC_ATAC', '09G_AACCATACGTTC_ATAC',
'09H_TGGCACAAAGGA_ATAC', '09H_GTTTACCTAGTT_ATAC',
'09F_TAGTTGGCAGAT_ATAC', '09L_CTTAGACGTCTA_ATAC',
'09C_GATGTAATTTGA_ATAC', '09I_TAGGCTTATTGT_ATAC'],
dtype='object', name='cells', length=9190))
这里我们将3000个细胞“去匿名”,以模拟部分配对数据集。
请不要在自己的数据上原样运行下面的代码块,您需要做的是保证自己的数据中配对细胞的
obs_name相同,不配对细胞的obs_name不同。
[5]:
rna_deanonym = rna.obs_names.str[:-4]
atac_deanonym = atac.obs_names.str[:-5]
assert set(rna_deanonym) == set(atac_deanonym) # The original barcodes are fully paired
paired_cells = set(Random(0).sample(set(rna_deanonym), 3000))
rna.obs_names = [
deanonym if deanonym in paired_cells else anonym
for anonym, deanonym in zip(rna.obs_names, rna_deanonym)
]
atac.obs_names = [
deanonym if deanonym in paired_cells else anonym
for anonym, deanonym in zip(atac.obs_names, atac_deanonym)
]
rna.obs_names, atac.obs_names
[5]:
(Index(['09L_GTTAGAACAGAT', '09I_CATAGCTCAACG_RNA', '09G_TTACAGGCAGGT',
'09B_CTATGGGCGTGC', '09L_AGTGATTGTTCG_RNA', '09C_ATATGTCTATAA_RNA',
'09A_CCTATTATTCGA_RNA', '09C_GCGTGCCGAATG_RNA', '09G_AGCTGACGCCGA',
'09D_CAGTCCCCAAAT_RNA',
...
'09H_GTTCGCTTGTCG_RNA', '09B_CAGGTGGTCTAT_RNA', '09C_GACCCTGTCGTT_RNA',
'09I_AACCAATCCTCA_RNA', '09C_AGAGTCTGAAAC_RNA', '09E_CCTGCTCGTCAC_RNA',
'09E_TGAGCACCTCCT_RNA', '09E_CCGGACTCGATC', '09F_GCCCGACTCGCA_RNA',
'09D_ATACTGCGCCCA_RNA'],
dtype='object', length=9190),
Index(['09L_GTAAACCCGATA_ATAC', '09D_GCCGGGCGGCCG_ATAC', '09J_TAGGCCTGAATT',
'09C_TCCGGCATCTCA_ATAC', '09C_AGGCCACGTACA', '09E_AAGGGGAGCTGG_ATAC',
'09K_CCGCCTTATAAT_ATAC', '09A_CGGATGTATCAT_ATAC', '09B_TCAGAACTGCTA',
'09J_TCCGTGATAGAG_ATAC',
...
'09E_TAGTCGCGTACT_ATAC', '09J_GCACCTAAATGA', '09E_CACCCACGCTCC',
'09G_AACCATACGTTC', '09H_TGGCACAAAGGA_ATAC', '09H_GTTTACCTAGTT',
'09F_TAGTTGGCAGAT_ATAC', '09L_CTTAGACGTCTA', '09C_GATGTAATTTGA_ATAC',
'09I_TAGGCTTATTGT'],
dtype='object', length=9190))
配置数据
在配置数据时,您需要在之前 第二步 设置的基础上额外添加 use_obs_names=True,以告诉模型使用 obs_names 来识别配对的细胞:
[6]:
scglue.models.configure_dataset(
rna, "NB", use_highly_variable=True,
use_layer="counts", use_rep="X_pca",
use_obs_names=True
)
[7]:
scglue.models.configure_dataset(
atac, "NB", use_highly_variable=True,
use_rep="X_lsi",
use_obs_names=True
)
[8]:
guidance_hvf = guidance.subgraph(chain(
rna.var.query("highly_variable").index,
atac.var.query("highly_variable").index
)).copy()
训练GLUE模型
我们仍然使用 scglue.models.fit_SCGLUE 函数来训练GLUE模型(参见 第二步),唯一的差别是要将 model 参数设置为 scglue.models.PairedSCGLUEModel:
[9]:
glue = scglue.models.fit_SCGLUE(
{"rna": rna, "atac": atac}, guidance_hvf,
model=scglue.models.PairedSCGLUEModel,
fit_kws={"directory": "glue"}
)
[INFO] fit_SCGLUE: Pretraining SCGLUE model...
[INFO] autodevice: Using GPU 1 as computation device.
[INFO] check_graph: Checking variable coverage...
[INFO] check_graph: Checking edge attributes...
[INFO] check_graph: Checking self-loops...
[INFO] check_graph: Checking graph symmetry...
[INFO] PairedSCGLUEModel: Setting `graph_batch_size` = 27025
[INFO] PairedSCGLUEModel: Setting `max_epochs` = 222
[INFO] PairedSCGLUEModel: Setting `patience` = 19
[INFO] PairedSCGLUEModel: Setting `reduce_lr_patience` = 10
[INFO] PairedSCGLUETrainer: Using training directory: "glue/pretrain"
[INFO] PairedSCGLUETrainer: [Epoch 10] train={'g_nll': 0.448, 'g_kl': 0.005, 'g_elbo': 0.452, 'x_rna_nll': 0.166, 'x_rna_kl': 0.007, 'x_rna_elbo': 0.173, 'x_atac_nll': 0.04, 'x_atac_kl': 0.001, 'x_atac_elbo': 0.041, 'dsc_loss': 0.692, 'vae_loss': 0.243, 'gen_loss': 0.209, 'joint_cross_loss': 0.206, 'real_cross_loss': 0.208, 'cos_loss': 0.131}, val={'g_nll': 0.445, 'g_kl': 0.005, 'g_elbo': 0.45, 'x_rna_nll': 0.168, 'x_rna_kl': 0.008, 'x_rna_elbo': 0.176, 'x_atac_nll': 0.041, 'x_atac_kl': 0.001, 'x_atac_elbo': 0.042, 'dsc_loss': 0.693, 'vae_loss': 0.247, 'gen_loss': 0.212, 'joint_cross_loss': 0.208, 'real_cross_loss': 0.209, 'cos_loss': 0.142}, 6.3s elapsed
[INFO] PairedSCGLUETrainer: [Epoch 20] train={'g_nll': 0.431, 'g_kl': 0.004, 'g_elbo': 0.434, 'x_rna_nll': 0.163, 'x_rna_kl': 0.007, 'x_rna_elbo': 0.17, 'x_atac_nll': 0.04, 'x_atac_kl': 0.001, 'x_atac_elbo': 0.04, 'dsc_loss': 0.693, 'vae_loss': 0.238, 'gen_loss': 0.203, 'joint_cross_loss': 0.202, 'real_cross_loss': 0.205, 'cos_loss': 0.131}, val={'g_nll': 0.43, 'g_kl': 0.004, 'g_elbo': 0.434, 'x_rna_nll': 0.165, 'x_rna_kl': 0.007, 'x_rna_elbo': 0.172, 'x_atac_nll': 0.041, 'x_atac_kl': 0.001, 'x_atac_elbo': 0.041, 'dsc_loss': 0.694, 'vae_loss': 0.241, 'gen_loss': 0.206, 'joint_cross_loss': 0.205, 'real_cross_loss': 0.205, 'cos_loss': 0.144}, 6.8s elapsed
[INFO] PairedSCGLUETrainer: [Epoch 30] train={'g_nll': 0.424, 'g_kl': 0.004, 'g_elbo': 0.427, 'x_rna_nll': 0.162, 'x_rna_kl': 0.007, 'x_rna_elbo': 0.168, 'x_atac_nll': 0.039, 'x_atac_kl': 0.001, 'x_atac_elbo': 0.04, 'dsc_loss': 0.693, 'vae_loss': 0.236, 'gen_loss': 0.202, 'joint_cross_loss': 0.201, 'real_cross_loss': 0.204, 'cos_loss': 0.131}, val={'g_nll': 0.424, 'g_kl': 0.004, 'g_elbo': 0.427, 'x_rna_nll': 0.162, 'x_rna_kl': 0.007, 'x_rna_elbo': 0.169, 'x_atac_nll': 0.04, 'x_atac_kl': 0.001, 'x_atac_elbo': 0.04, 'dsc_loss': 0.694, 'vae_loss': 0.237, 'gen_loss': 0.202, 'joint_cross_loss': 0.204, 'real_cross_loss': 0.206, 'cos_loss': 0.147}, 6.6s elapsed
[INFO] PairedSCGLUETrainer: [Epoch 40] train={'g_nll': 0.42, 'g_kl': 0.004, 'g_elbo': 0.424, 'x_rna_nll': 0.16, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.167, 'x_atac_nll': 0.039, 'x_atac_kl': 0.001, 'x_atac_elbo': 0.04, 'dsc_loss': 0.693, 'vae_loss': 0.234, 'gen_loss': 0.2, 'joint_cross_loss': 0.201, 'real_cross_loss': 0.204, 'cos_loss': 0.131}, val={'g_nll': 0.419, 'g_kl': 0.004, 'g_elbo': 0.423, 'x_rna_nll': 0.166, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.172, 'x_atac_nll': 0.04, 'x_atac_kl': 0.001, 'x_atac_elbo': 0.04, 'dsc_loss': 0.694, 'vae_loss': 0.24, 'gen_loss': 0.206, 'joint_cross_loss': 0.204, 'real_cross_loss': 0.205, 'cos_loss': 0.153}, 6.7s elapsed
Epoch 00041: reducing learning rate of group 0 to 2.0000e-04.
Epoch 00041: reducing learning rate of group 0 to 2.0000e-04.
[INFO] LRScheduler: Learning rate reduction: step 1
[INFO] PairedSCGLUETrainer: [Epoch 50] train={'g_nll': 0.418, 'g_kl': 0.004, 'g_elbo': 0.421, 'x_rna_nll': 0.16, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.167, 'x_atac_nll': 0.039, 'x_atac_kl': 0.001, 'x_atac_elbo': 0.04, 'dsc_loss': 0.693, 'vae_loss': 0.234, 'gen_loss': 0.199, 'joint_cross_loss': 0.199, 'real_cross_loss': 0.203, 'cos_loss': 0.128}, val={'g_nll': 0.418, 'g_kl': 0.004, 'g_elbo': 0.422, 'x_rna_nll': 0.161, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.168, 'x_atac_nll': 0.041, 'x_atac_kl': 0.001, 'x_atac_elbo': 0.042, 'dsc_loss': 0.693, 'vae_loss': 0.237, 'gen_loss': 0.203, 'joint_cross_loss': 0.202, 'real_cross_loss': 0.205, 'cos_loss': 0.147}, 6.2s elapsed
[INFO] PairedSCGLUETrainer: [Epoch 60] train={'g_nll': 0.418, 'g_kl': 0.004, 'g_elbo': 0.421, 'x_rna_nll': 0.161, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.167, 'x_atac_nll': 0.039, 'x_atac_kl': 0.001, 'x_atac_elbo': 0.04, 'dsc_loss': 0.693, 'vae_loss': 0.234, 'gen_loss': 0.2, 'joint_cross_loss': 0.199, 'real_cross_loss': 0.202, 'cos_loss': 0.128}, val={'g_nll': 0.417, 'g_kl': 0.004, 'g_elbo': 0.421, 'x_rna_nll': 0.161, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.167, 'x_atac_nll': 0.04, 'x_atac_kl': 0.001, 'x_atac_elbo': 0.041, 'dsc_loss': 0.693, 'vae_loss': 0.236, 'gen_loss': 0.201, 'joint_cross_loss': 0.202, 'real_cross_loss': 0.204, 'cos_loss': 0.15}, 6.9s elapsed
Epoch 00067: reducing learning rate of group 0 to 2.0000e-05.
Epoch 00067: reducing learning rate of group 0 to 2.0000e-05.
[INFO] LRScheduler: Learning rate reduction: step 2
[INFO] PairedSCGLUETrainer: [Epoch 70] train={'g_nll': 0.417, 'g_kl': 0.004, 'g_elbo': 0.421, 'x_rna_nll': 0.161, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.167, 'x_atac_nll': 0.039, 'x_atac_kl': 0.001, 'x_atac_elbo': 0.04, 'dsc_loss': 0.693, 'vae_loss': 0.234, 'gen_loss': 0.2, 'joint_cross_loss': 0.199, 'real_cross_loss': 0.203, 'cos_loss': 0.127}, val={'g_nll': 0.418, 'g_kl': 0.004, 'g_elbo': 0.421, 'x_rna_nll': 0.161, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.167, 'x_atac_nll': 0.041, 'x_atac_kl': 0.001, 'x_atac_elbo': 0.041, 'dsc_loss': 0.693, 'vae_loss': 0.236, 'gen_loss': 0.202, 'joint_cross_loss': 0.203, 'real_cross_loss': 0.205, 'cos_loss': 0.153}, 6.2s elapsed
2022-08-26 16:36:30,211 ignite.handlers.early_stopping.EarlyStopping INFO: EarlyStopping: Stop training
[INFO] EarlyStopping: Restoring checkpoint "69"...
[INFO] fit_SCGLUE: Estimating balancing weight...
[INFO] estimate_balancing_weight: Clustering cells...
[INFO] estimate_balancing_weight: Matching clusters...
[INFO] estimate_balancing_weight: Matching array shape = (16, 20)...
[INFO] estimate_balancing_weight: Estimating balancing weight...
[INFO] fit_SCGLUE: Fine-tuning SCGLUE model...
[INFO] check_graph: Checking variable coverage...
[INFO] check_graph: Checking edge attributes...
[INFO] check_graph: Checking self-loops...
[INFO] check_graph: Checking graph symmetry...
[INFO] PairedSCGLUEModel: Setting `graph_batch_size` = 27025
[INFO] PairedSCGLUEModel: Setting `align_burnin` = 37
[INFO] PairedSCGLUEModel: Setting `max_epochs` = 222
[INFO] PairedSCGLUEModel: Setting `patience` = 19
[INFO] PairedSCGLUEModel: Setting `reduce_lr_patience` = 10
[INFO] PairedSCGLUETrainer: Using training directory: "glue/fine-tune"
[INFO] PairedSCGLUETrainer: [Epoch 10] train={'g_nll': 0.416, 'g_kl': 0.004, 'g_elbo': 0.419, 'x_rna_nll': 0.161, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.167, 'x_atac_nll': 0.039, 'x_atac_kl': 0.001, 'x_atac_elbo': 0.04, 'dsc_loss': 0.688, 'vae_loss': 0.234, 'gen_loss': 0.2, 'joint_cross_loss': 0.2, 'real_cross_loss': 0.203, 'cos_loss': 0.133}, val={'g_nll': 0.414, 'g_kl': 0.004, 'g_elbo': 0.418, 'x_rna_nll': 0.161, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.167, 'x_atac_nll': 0.04, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.041, 'dsc_loss': 0.702, 'vae_loss': 0.235, 'gen_loss': 0.2, 'joint_cross_loss': 0.202, 'real_cross_loss': 0.206, 'cos_loss': 0.143}, 6.7s elapsed
[INFO] PairedSCGLUETrainer: [Epoch 20] train={'g_nll': 0.413, 'g_kl': 0.004, 'g_elbo': 0.417, 'x_rna_nll': 0.16, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.166, 'x_atac_nll': 0.039, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.04, 'dsc_loss': 0.69, 'vae_loss': 0.234, 'gen_loss': 0.199, 'joint_cross_loss': 0.2, 'real_cross_loss': 0.203, 'cos_loss': 0.134}, val={'g_nll': 0.414, 'g_kl': 0.004, 'g_elbo': 0.417, 'x_rna_nll': 0.163, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.169, 'x_atac_nll': 0.041, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.041, 'dsc_loss': 0.714, 'vae_loss': 0.238, 'gen_loss': 0.202, 'joint_cross_loss': 0.205, 'real_cross_loss': 0.206, 'cos_loss': 0.141}, 6.5s elapsed
[INFO] PairedSCGLUETrainer: [Epoch 30] train={'g_nll': 0.411, 'g_kl': 0.004, 'g_elbo': 0.415, 'x_rna_nll': 0.161, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.167, 'x_atac_nll': 0.039, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.04, 'dsc_loss': 0.689, 'vae_loss': 0.234, 'gen_loss': 0.199, 'joint_cross_loss': 0.2, 'real_cross_loss': 0.203, 'cos_loss': 0.135}, val={'g_nll': 0.412, 'g_kl': 0.004, 'g_elbo': 0.415, 'x_rna_nll': 0.163, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.169, 'x_atac_nll': 0.041, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.041, 'dsc_loss': 0.71, 'vae_loss': 0.238, 'gen_loss': 0.202, 'joint_cross_loss': 0.204, 'real_cross_loss': 0.206, 'cos_loss': 0.149}, 6.6s elapsed
[INFO] PairedSCGLUETrainer: [Epoch 40] train={'g_nll': 0.41, 'g_kl': 0.004, 'g_elbo': 0.413, 'x_rna_nll': 0.16, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.166, 'x_atac_nll': 0.039, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.04, 'dsc_loss': 0.694, 'vae_loss': 0.233, 'gen_loss': 0.198, 'joint_cross_loss': 0.2, 'real_cross_loss': 0.203, 'cos_loss': 0.135}, val={'g_nll': 0.41, 'g_kl': 0.004, 'g_elbo': 0.413, 'x_rna_nll': 0.164, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.17, 'x_atac_nll': 0.04, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.041, 'dsc_loss': 0.713, 'vae_loss': 0.239, 'gen_loss': 0.203, 'joint_cross_loss': 0.204, 'real_cross_loss': 0.204, 'cos_loss': 0.147}, 6.4s elapsed
[INFO] PairedSCGLUETrainer: [Epoch 50] train={'g_nll': 0.409, 'g_kl': 0.004, 'g_elbo': 0.412, 'x_rna_nll': 0.161, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.166, 'x_atac_nll': 0.039, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.04, 'dsc_loss': 0.689, 'vae_loss': 0.233, 'gen_loss': 0.199, 'joint_cross_loss': 0.199, 'real_cross_loss': 0.202, 'cos_loss': 0.136}, val={'g_nll': 0.409, 'g_kl': 0.004, 'g_elbo': 0.412, 'x_rna_nll': 0.164, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.17, 'x_atac_nll': 0.041, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.041, 'dsc_loss': 0.693, 'vae_loss': 0.239, 'gen_loss': 0.204, 'joint_cross_loss': 0.205, 'real_cross_loss': 0.207, 'cos_loss': 0.15}, 6.6s elapsed
[INFO] PairedSCGLUETrainer: [Epoch 60] train={'g_nll': 0.409, 'g_kl': 0.004, 'g_elbo': 0.412, 'x_rna_nll': 0.16, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.166, 'x_atac_nll': 0.039, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.04, 'dsc_loss': 0.692, 'vae_loss': 0.233, 'gen_loss': 0.198, 'joint_cross_loss': 0.199, 'real_cross_loss': 0.202, 'cos_loss': 0.135}, val={'g_nll': 0.409, 'g_kl': 0.004, 'g_elbo': 0.412, 'x_rna_nll': 0.163, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.169, 'x_atac_nll': 0.04, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.041, 'dsc_loss': 0.709, 'vae_loss': 0.238, 'gen_loss': 0.202, 'joint_cross_loss': 0.203, 'real_cross_loss': 0.206, 'cos_loss': 0.15}, 6.4s elapsed
[INFO] PairedSCGLUETrainer: [Epoch 70] train={'g_nll': 0.408, 'g_kl': 0.004, 'g_elbo': 0.412, 'x_rna_nll': 0.16, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.165, 'x_atac_nll': 0.039, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.04, 'dsc_loss': 0.687, 'vae_loss': 0.232, 'gen_loss': 0.198, 'joint_cross_loss': 0.199, 'real_cross_loss': 0.202, 'cos_loss': 0.136}, val={'g_nll': 0.409, 'g_kl': 0.003, 'g_elbo': 0.412, 'x_rna_nll': 0.16, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.166, 'x_atac_nll': 0.04, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.041, 'dsc_loss': 0.715, 'vae_loss': 0.234, 'gen_loss': 0.199, 'joint_cross_loss': 0.201, 'real_cross_loss': 0.203, 'cos_loss': 0.151}, 6.6s elapsed
[INFO] PairedSCGLUETrainer: [Epoch 80] train={'g_nll': 0.408, 'g_kl': 0.004, 'g_elbo': 0.412, 'x_rna_nll': 0.16, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.166, 'x_atac_nll': 0.039, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.04, 'dsc_loss': 0.692, 'vae_loss': 0.232, 'gen_loss': 0.198, 'joint_cross_loss': 0.199, 'real_cross_loss': 0.202, 'cos_loss': 0.135}, val={'g_nll': 0.408, 'g_kl': 0.004, 'g_elbo': 0.411, 'x_rna_nll': 0.164, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.17, 'x_atac_nll': 0.04, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.041, 'dsc_loss': 0.707, 'vae_loss': 0.238, 'gen_loss': 0.203, 'joint_cross_loss': 0.203, 'real_cross_loss': 0.206, 'cos_loss': 0.152}, 6.5s elapsed
Epoch 00081: reducing learning rate of group 0 to 2.0000e-04.
Epoch 00081: reducing learning rate of group 0 to 2.0000e-04.
[INFO] LRScheduler: Learning rate reduction: step 1
[INFO] PairedSCGLUETrainer: [Epoch 90] train={'g_nll': 0.407, 'g_kl': 0.003, 'g_elbo': 0.411, 'x_rna_nll': 0.16, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.165, 'x_atac_nll': 0.039, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.039, 'dsc_loss': 0.692, 'vae_loss': 0.232, 'gen_loss': 0.197, 'joint_cross_loss': 0.199, 'real_cross_loss': 0.201, 'cos_loss': 0.133}, val={'g_nll': 0.407, 'g_kl': 0.003, 'g_elbo': 0.41, 'x_rna_nll': 0.164, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.17, 'x_atac_nll': 0.041, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.041, 'dsc_loss': 0.708, 'vae_loss': 0.239, 'gen_loss': 0.203, 'joint_cross_loss': 0.206, 'real_cross_loss': 0.208, 'cos_loss': 0.148}, 6.7s elapsed
Epoch 00092: reducing learning rate of group 0 to 2.0000e-05.
Epoch 00092: reducing learning rate of group 0 to 2.0000e-05.
[INFO] LRScheduler: Learning rate reduction: step 2
[INFO] PairedSCGLUETrainer: [Epoch 100] train={'g_nll': 0.407, 'g_kl': 0.003, 'g_elbo': 0.41, 'x_rna_nll': 0.16, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.166, 'x_atac_nll': 0.039, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.04, 'dsc_loss': 0.689, 'vae_loss': 0.232, 'gen_loss': 0.198, 'joint_cross_loss': 0.199, 'real_cross_loss': 0.201, 'cos_loss': 0.133}, val={'g_nll': 0.407, 'g_kl': 0.003, 'g_elbo': 0.41, 'x_rna_nll': 0.161, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.167, 'x_atac_nll': 0.04, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.041, 'dsc_loss': 0.709, 'vae_loss': 0.235, 'gen_loss': 0.199, 'joint_cross_loss': 0.203, 'real_cross_loss': 0.207, 'cos_loss': 0.146}, 6.9s elapsed
Epoch 00103: reducing learning rate of group 0 to 2.0000e-06.
Epoch 00103: reducing learning rate of group 0 to 2.0000e-06.
[INFO] LRScheduler: Learning rate reduction: step 3
[INFO] PairedSCGLUETrainer: [Epoch 110] train={'g_nll': 0.407, 'g_kl': 0.003, 'g_elbo': 0.41, 'x_rna_nll': 0.16, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.166, 'x_atac_nll': 0.039, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.04, 'dsc_loss': 0.69, 'vae_loss': 0.232, 'gen_loss': 0.198, 'joint_cross_loss': 0.199, 'real_cross_loss': 0.201, 'cos_loss': 0.134}, val={'g_nll': 0.407, 'g_kl': 0.003, 'g_elbo': 0.41, 'x_rna_nll': 0.163, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.168, 'x_atac_nll': 0.04, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.04, 'dsc_loss': 0.712, 'vae_loss': 0.236, 'gen_loss': 0.201, 'joint_cross_loss': 0.203, 'real_cross_loss': 0.206, 'cos_loss': 0.151}, 8.4s elapsed
Epoch 00114: reducing learning rate of group 0 to 2.0000e-07.
Epoch 00114: reducing learning rate of group 0 to 2.0000e-07.
[INFO] LRScheduler: Learning rate reduction: step 4
[INFO] PairedSCGLUETrainer: [Epoch 120] train={'g_nll': 0.407, 'g_kl': 0.003, 'g_elbo': 0.41, 'x_rna_nll': 0.16, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.165, 'x_atac_nll': 0.039, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.04, 'dsc_loss': 0.689, 'vae_loss': 0.232, 'gen_loss': 0.198, 'joint_cross_loss': 0.199, 'real_cross_loss': 0.201, 'cos_loss': 0.133}, val={'g_nll': 0.407, 'g_kl': 0.003, 'g_elbo': 0.411, 'x_rna_nll': 0.162, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.168, 'x_atac_nll': 0.04, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.041, 'dsc_loss': 0.708, 'vae_loss': 0.236, 'gen_loss': 0.201, 'joint_cross_loss': 0.203, 'real_cross_loss': 0.205, 'cos_loss': 0.149}, 8.3s elapsed
Epoch 00128: reducing learning rate of group 0 to 2.0000e-08.
Epoch 00128: reducing learning rate of group 0 to 2.0000e-08.
[INFO] LRScheduler: Learning rate reduction: step 5
[INFO] PairedSCGLUETrainer: [Epoch 130] train={'g_nll': 0.407, 'g_kl': 0.003, 'g_elbo': 0.41, 'x_rna_nll': 0.16, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.166, 'x_atac_nll': 0.039, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.04, 'dsc_loss': 0.688, 'vae_loss': 0.232, 'gen_loss': 0.198, 'joint_cross_loss': 0.199, 'real_cross_loss': 0.201, 'cos_loss': 0.133}, val={'g_nll': 0.407, 'g_kl': 0.003, 'g_elbo': 0.411, 'x_rna_nll': 0.161, 'x_rna_kl': 0.006, 'x_rna_elbo': 0.167, 'x_atac_nll': 0.041, 'x_atac_kl': 0.0, 'x_atac_elbo': 0.042, 'dsc_loss': 0.716, 'vae_loss': 0.236, 'gen_loss': 0.2, 'joint_cross_loss': 0.202, 'real_cross_loss': 0.203, 'cos_loss': 0.146}, 7.0s elapsed
2022-08-26 16:54:00,388 ignite.handlers.early_stopping.EarlyStopping INFO: EarlyStopping: Stop training
[INFO] EarlyStopping: Restoring checkpoint "122"...
结果模型将是一个 scglue.models.PairedSCGLUEModel 对象,但所有功能仍然保持不变。
继续参照 第二步 进行
从这里开始,您可以继续参照 第二步 的说明来获取细胞和特征嵌入:
[10]:
dx = scglue.models.integration_consistency(
glue, {"rna": rna, "atac": atac}, guidance_hvf
)
dx
[INFO] integration_consistency: Using layer "counts" for modality "rna"
[INFO] integration_consistency: Selecting aggregation "sum" for modality "rna"
[INFO] integration_consistency: Selecting aggregation "sum" for modality "atac"
[INFO] integration_consistency: Selecting log-norm preprocessing for modality "rna"
[INFO] integration_consistency: Selecting log-norm preprocessing for modality "atac"
[INFO] get_metacells: Clustering metacells...
[INFO] get_metacells: Aggregating metacells...
[INFO] metacell_corr: Computing correlation on 10 common metacells...
[INFO] get_metacells: Clustering metacells...
[INFO] get_metacells: Aggregating metacells...
[INFO] metacell_corr: Computing correlation on 20 common metacells...
[INFO] get_metacells: Clustering metacells...
[INFO] get_metacells: Aggregating metacells...
[INFO] metacell_corr: Computing correlation on 50 common metacells...
[INFO] get_metacells: Clustering metacells...
[INFO] get_metacells: Aggregating metacells...
[INFO] metacell_corr: Computing correlation on 100 common metacells...
[INFO] get_metacells: Clustering metacells...
[INFO] get_metacells: Aggregating metacells...
[INFO] metacell_corr: Computing correlation on 200 common metacells...
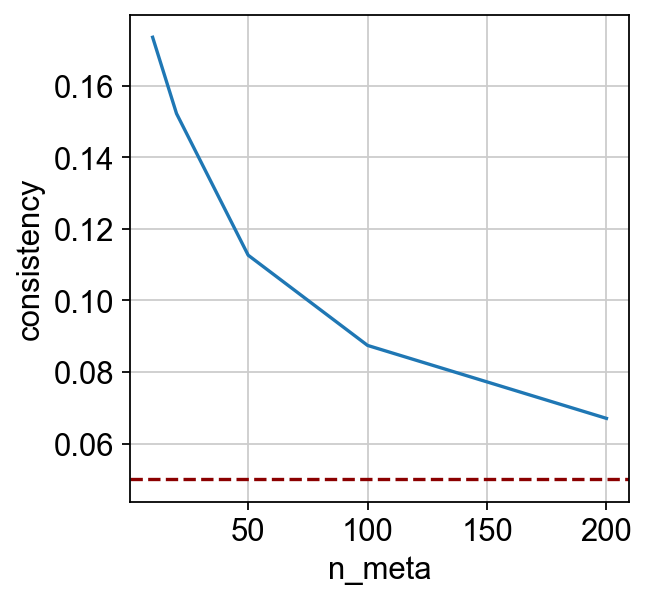
[10]:
| n_meta | consistency | |
|---|---|---|
| 0 | 10 | 0.173545 |
| 1 | 20 | 0.152228 |
| 2 | 50 | 0.112656 |
| 3 | 100 | 0.087442 |
| 4 | 200 | 0.067064 |
[11]:
_ = sns.lineplot(x="n_meta", y="consistency", data=dx).axhline(y=0.05, c="darkred", ls="--")
[12]:
rna.obsm["X_glue"] = glue.encode_data("rna", rna)
atac.obsm["X_glue"] = glue.encode_data("atac", atac)
[13]:
combined = ad.concat([rna, atac])
/rd2/user/caozj/GLUE/conda/lib/python3.8/site-packages/anndata/_core/anndata.py:1828: UserWarning: Observation names are not unique. To make them unique, call `.obs_names_make_unique`.
utils.warn_names_duplicates("obs")
[14]:
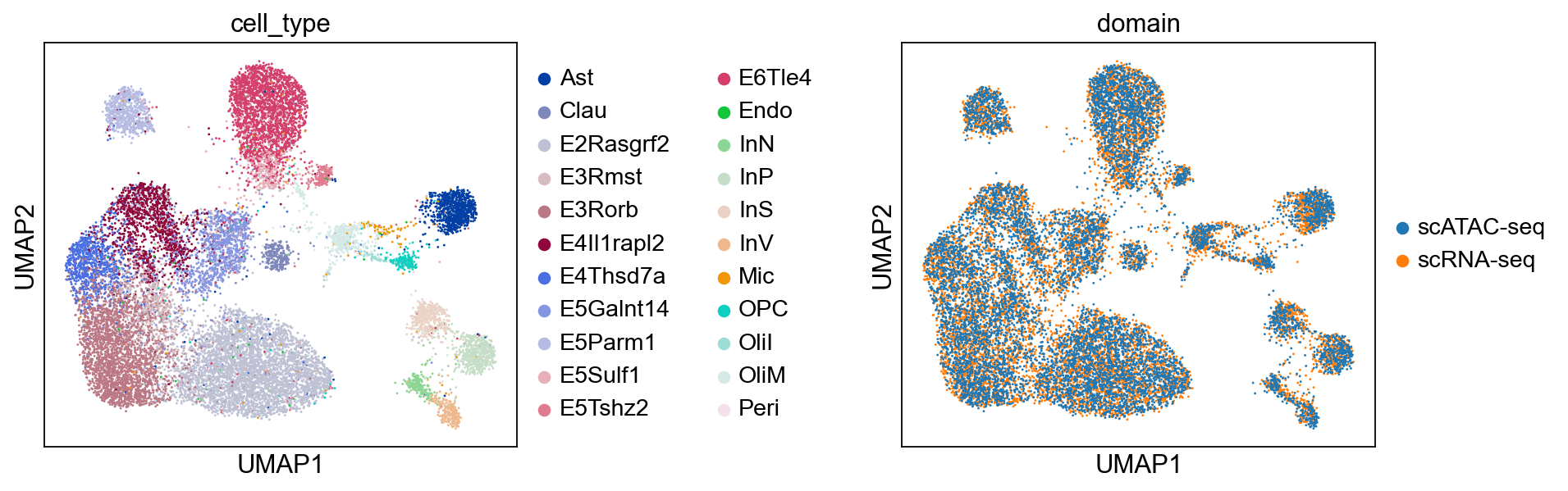
sc.pp.neighbors(combined, use_rep="X_glue", metric="cosine")
sc.tl.umap(combined)
sc.pl.umap(combined, color=["cell_type", "domain"], wspace=0.65)
[15]:
feature_embeddings = glue.encode_graph(guidance_hvf)
feature_embeddings = pd.DataFrame(feature_embeddings, index=glue.vertices)
feature_embeddings.iloc[:5, :5]
[15]:
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| 0610009B22Rik | -0.003289 | -0.101665 | 0.006089 | -0.005999 | -0.002966 |
| 0610025J13Rik | -0.002636 | 0.180847 | 0.000731 | 0.001941 | -0.006124 |
| 1110002J07Rik | -0.010751 | 0.079403 | 0.012048 | -0.011231 | -0.007835 |
| 1110006O24Rik | 0.003695 | -0.352514 | 0.000812 | -0.001779 | -0.003719 |
| 1110020A21Rik | -0.011558 | -0.251546 | 0.008463 | -0.003183 | -0.006828 |
[16]:
rna.varm["X_glue"] = feature_embeddings.reindex(rna.var_names).to_numpy()
atac.varm["X_glue"] = feature_embeddings.reindex(atac.var_names).to_numpy()